Navigation
What is DrugCVar
Targeted therapy for cancer-related genetic variants is critical for precision medicine, and next-generation sequencing based discovery of genetic variants become popular in cancer studies and clinical practice. Although several databases including CIViC, OncoKB, CGI, MCG provide clinical interpretations of variants in cancer, the clinical evidence was limited and miscellaneous.
In our study, we provide the DrugCVar database, which integrated our manually curated cancer variant-drug targeting evidences from literature and the interpretations from the public resources. In total, 8,539 clinical evidences for cancer variant-drug targeting were integrated and classified into ten evidence tiers. Searching and browsing functions were provided for quick queries of cancer variant-drug targeting evidence. Also, batch annotation was developed for use-provided massive genetic variants in various formats. Details such as the mutation function, location of the variants in gene and protein structures, and mutation statistics of queried genes in various tumor types were also provided for further investigations.
Thus, DrugCVar could serve as a comprehensive annotation tool to interpret potential drugs for cancer variants especially the massive ones from clinical cancer genomics studies.
How to use DrugCVar
Search
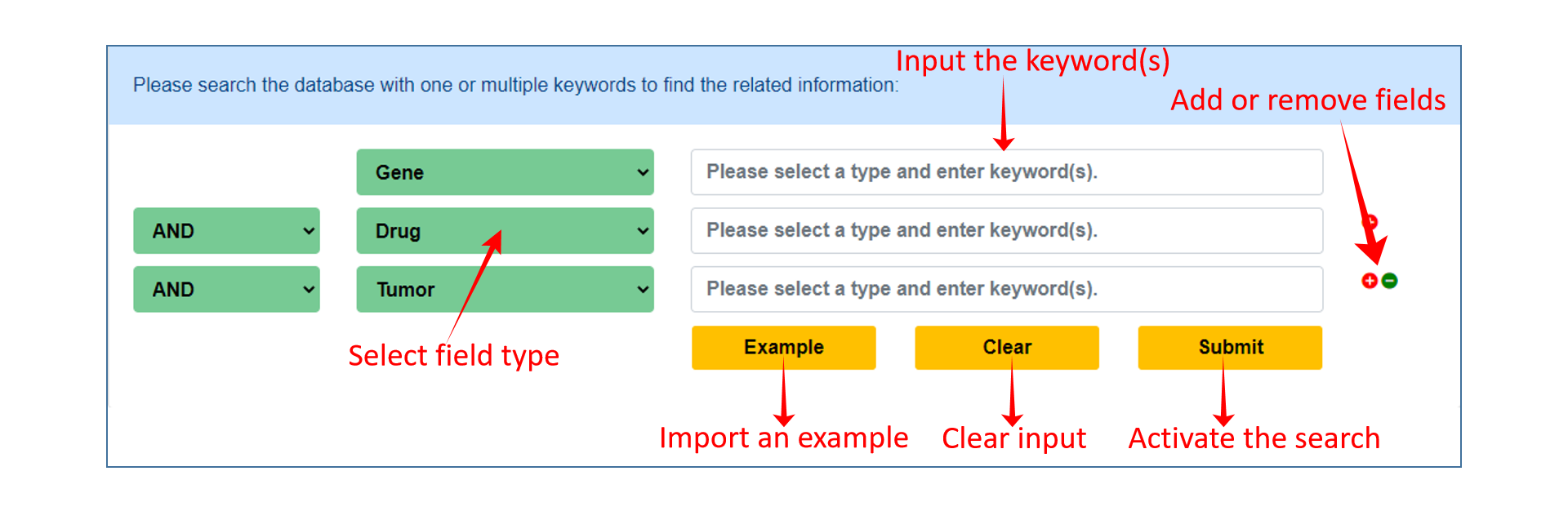
On the Search page of DrugCVar, users can specify one or more fields to perform a quick search with the keyword(s) in the field(s). Users can simply click example and submit buttons to perform an example search.
Browse
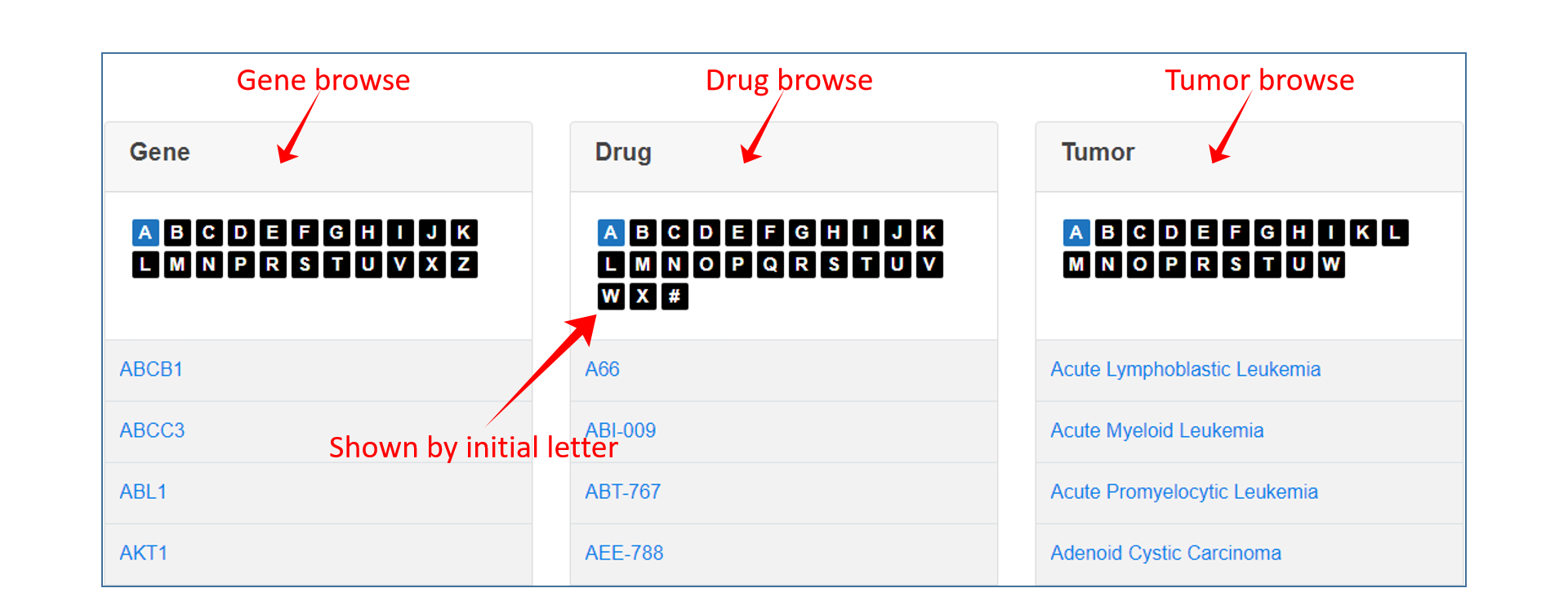
On the Browse page, users can click one term to view the specified data in DrugCVar. The browse page contains three modules including Gene, Drug and Tumor, which are sorted alphabetically. Users can click the initial letter and select the term of interest.
Search&Browse result
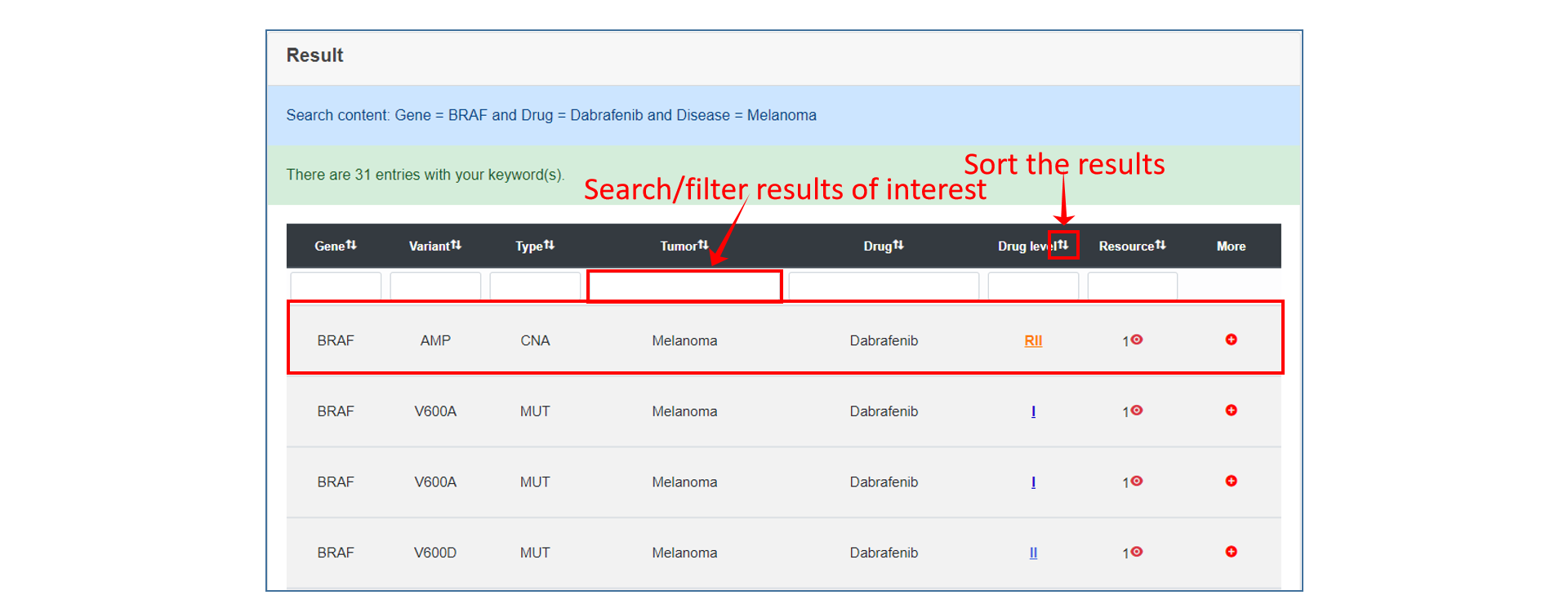
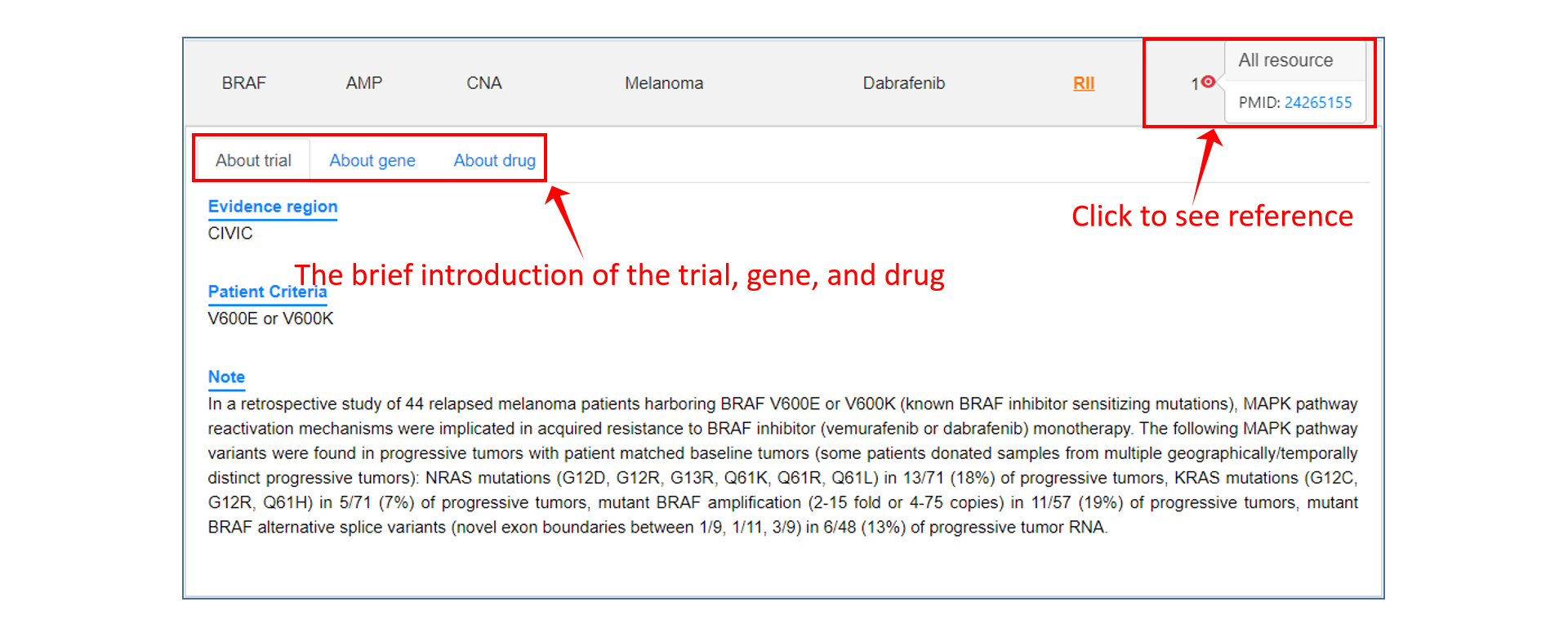
On the result page, users can view the search or browse results, and further sort/filter them by the options we provide.
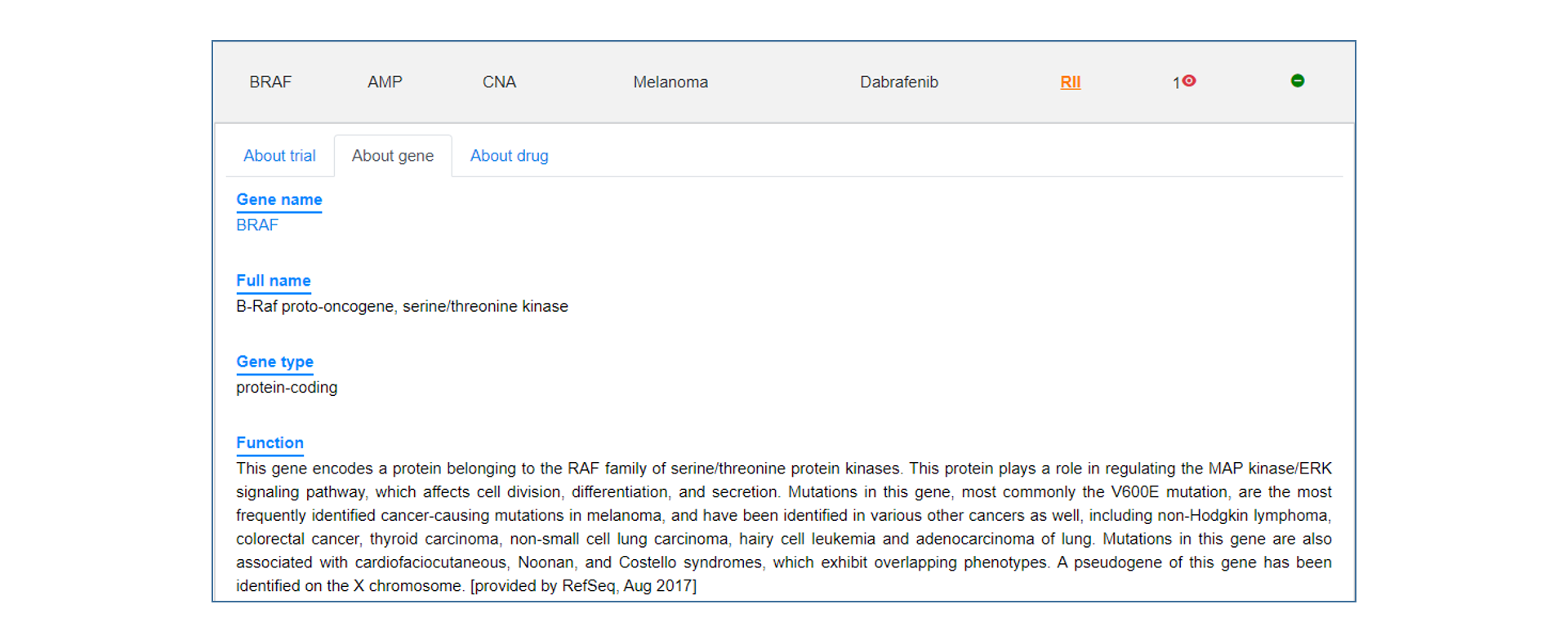
Analysis
Analysis function is mainly for in-depth analysis of tumor-related mutations. DrugCVar supports the function and drug annotation of single or batch mutations. Users can enter quarry terms or upload a file in a specified format of mutations for annotation analysis. Specifically, users need to do the following:
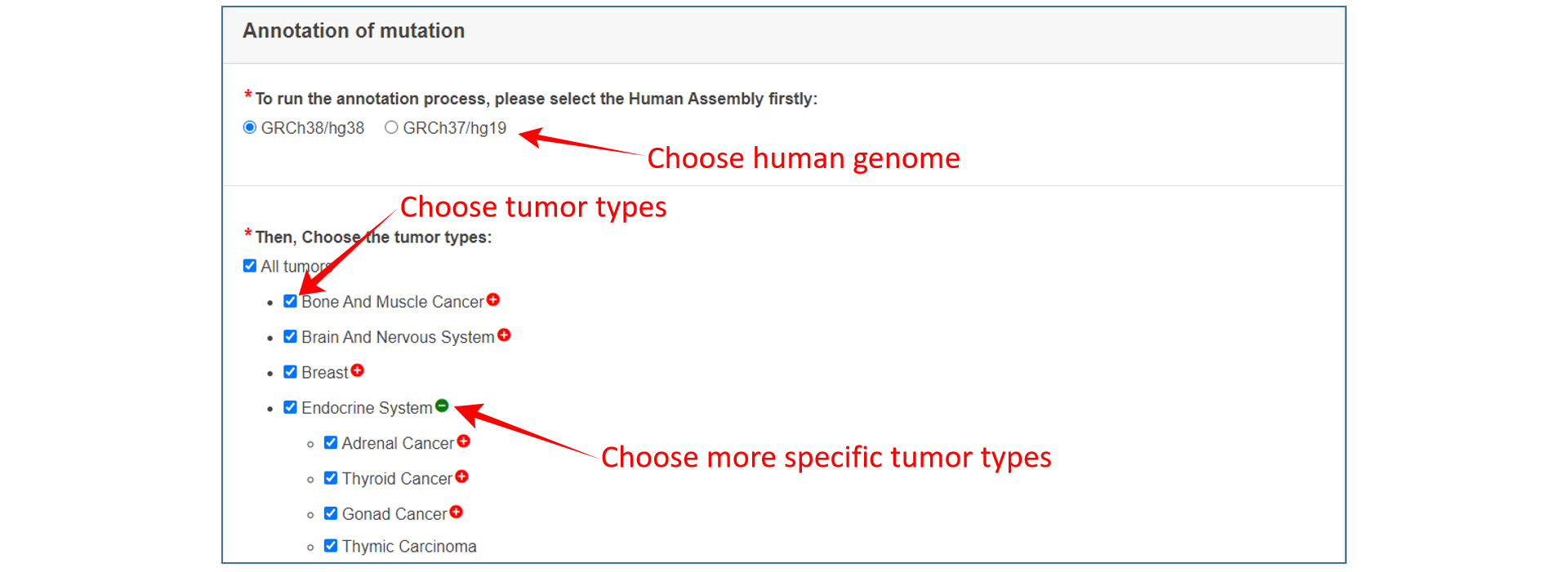
1、 Firstly, select the version of reference genome.
2、 Secondly, choose interested tumor type(s).
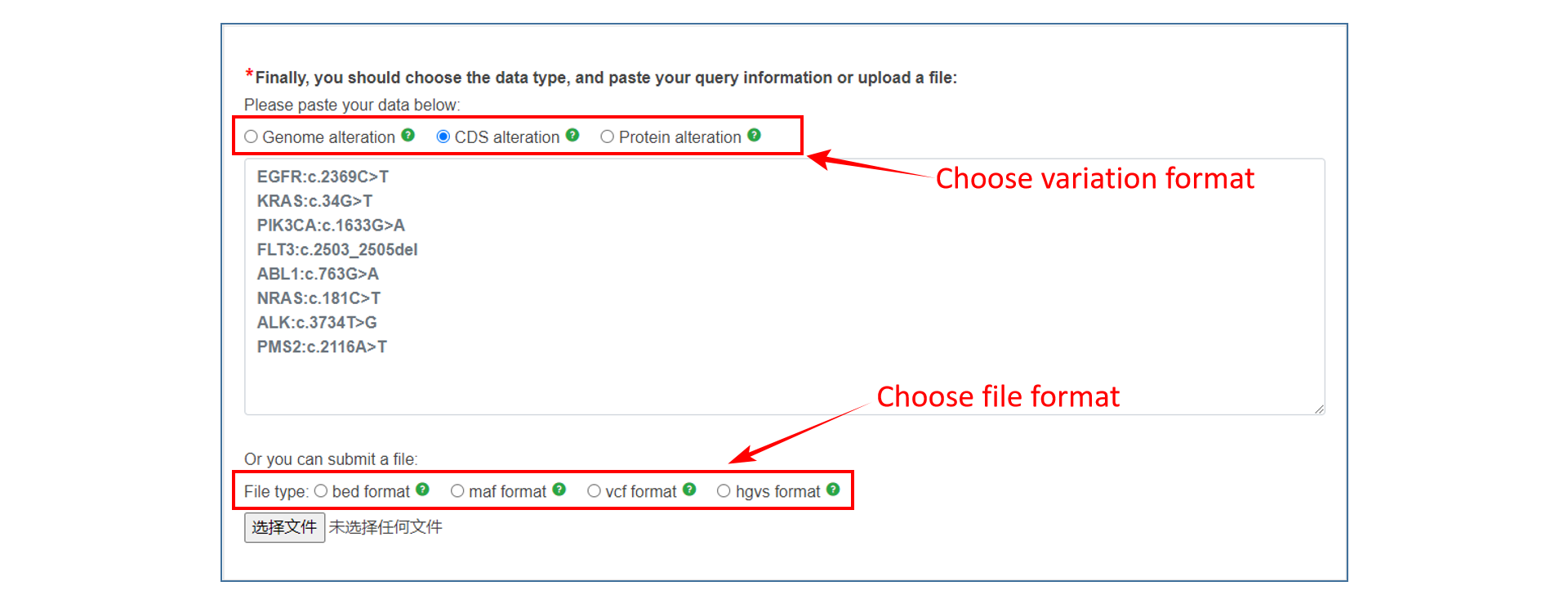
3、 Select mutation format and enter mutation terms manually or upload a corresponding file.
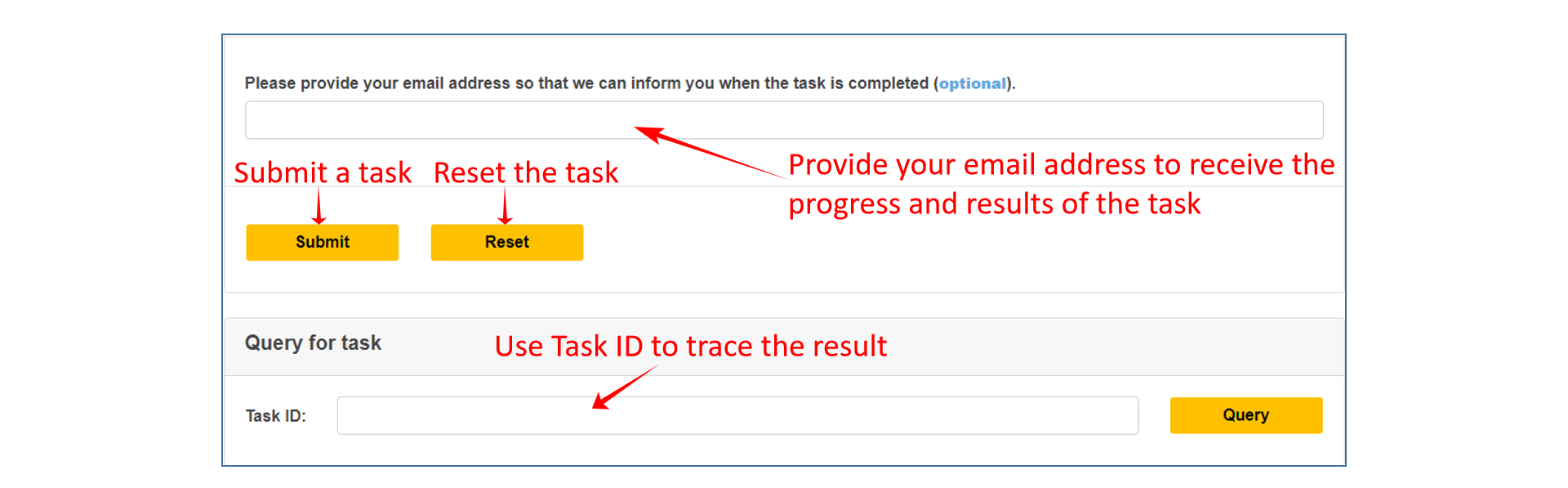
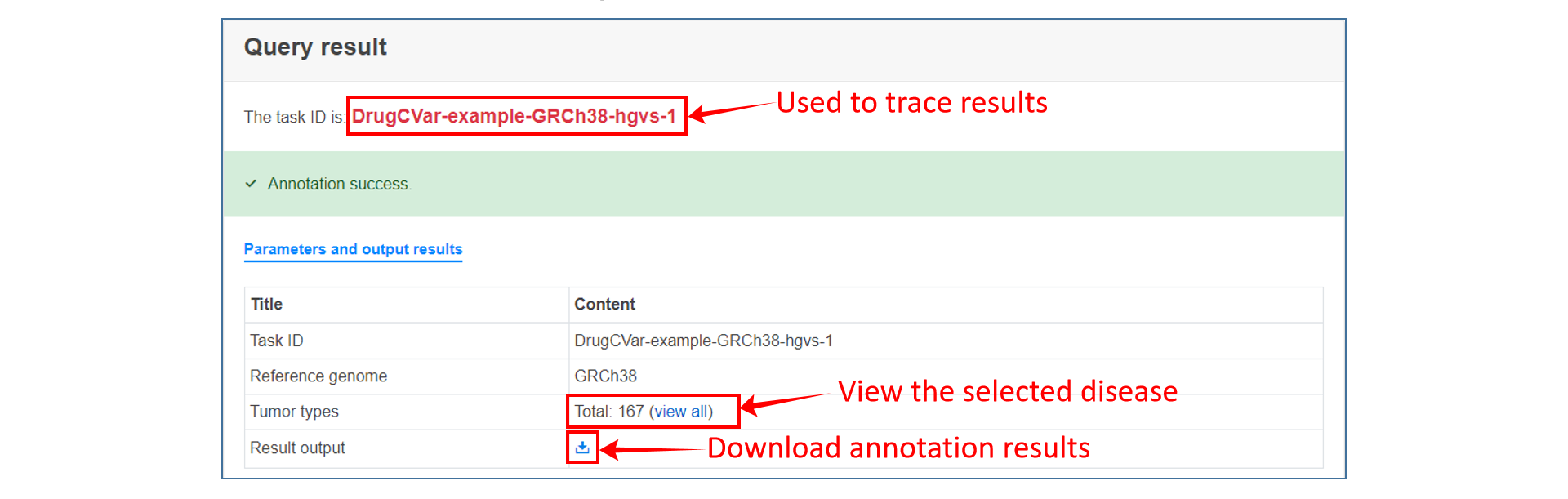
4、 Submit the task. If there are many mutations submitted, it may take some time. Users can optionally provide an email address and let the task run offline, and the Task ID of the final result will be sent to the user via email. Users can trace the task results through Task ID.
Analysis result
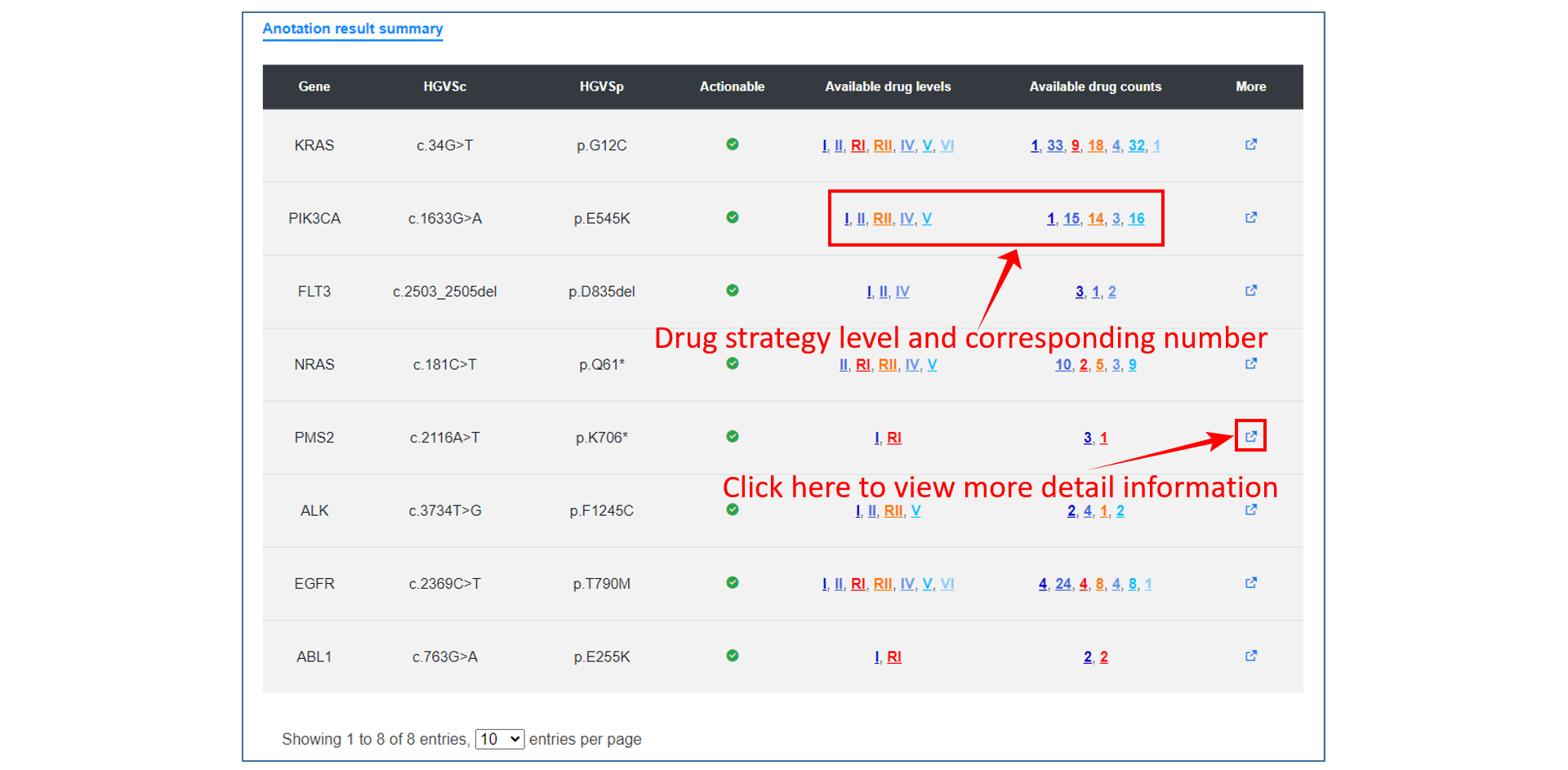
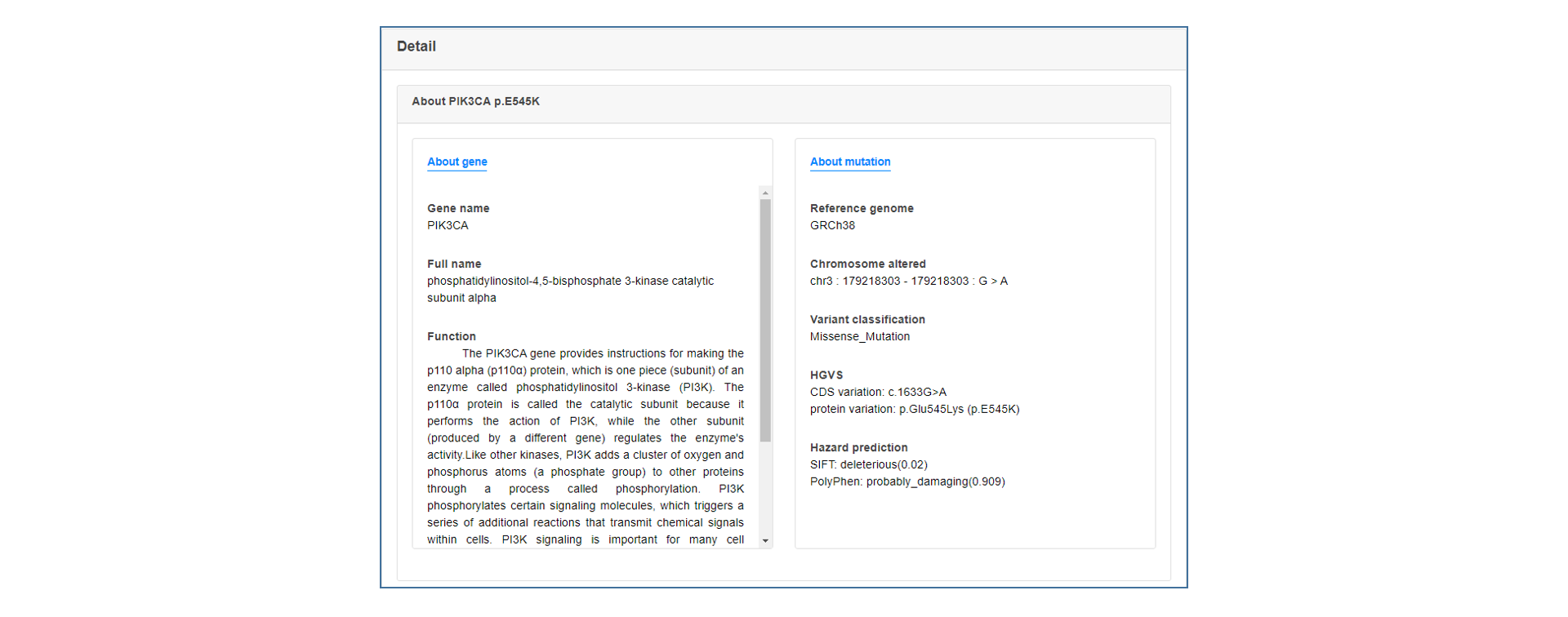
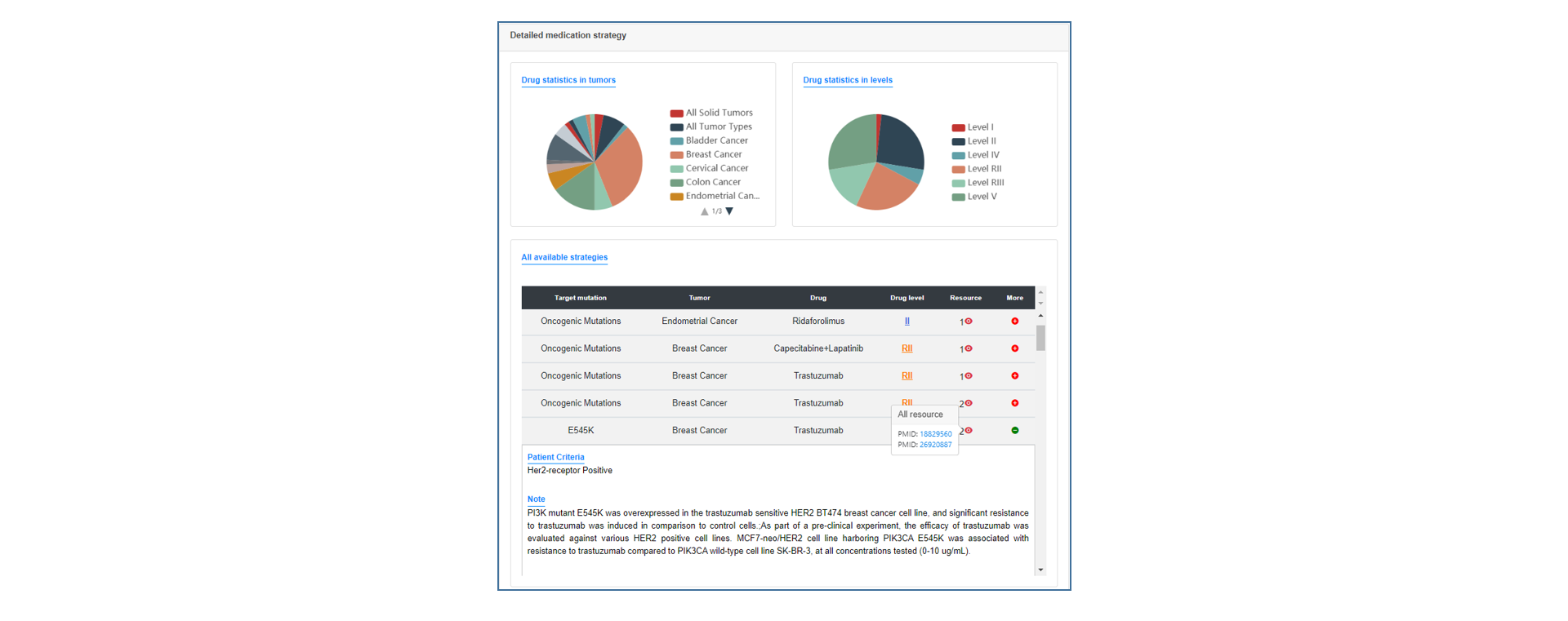
On the annotation result page, users can see the summary table of mutation annotations, including CDS change and protein change of gene and matched target drug information. users can click the "More" button to get a more detailed annotation result of each item, which contains introduction to gene and mutation function, medication strategy, mutation visualization and mutation background in TCGA.
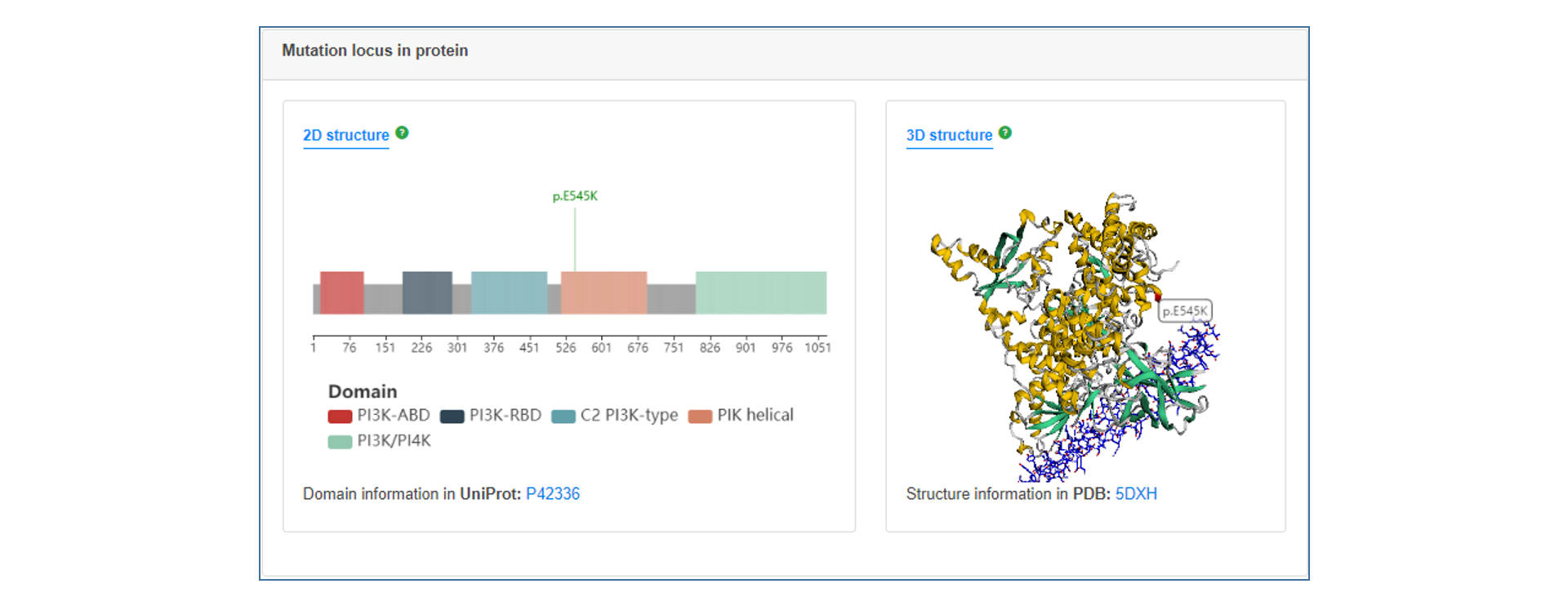
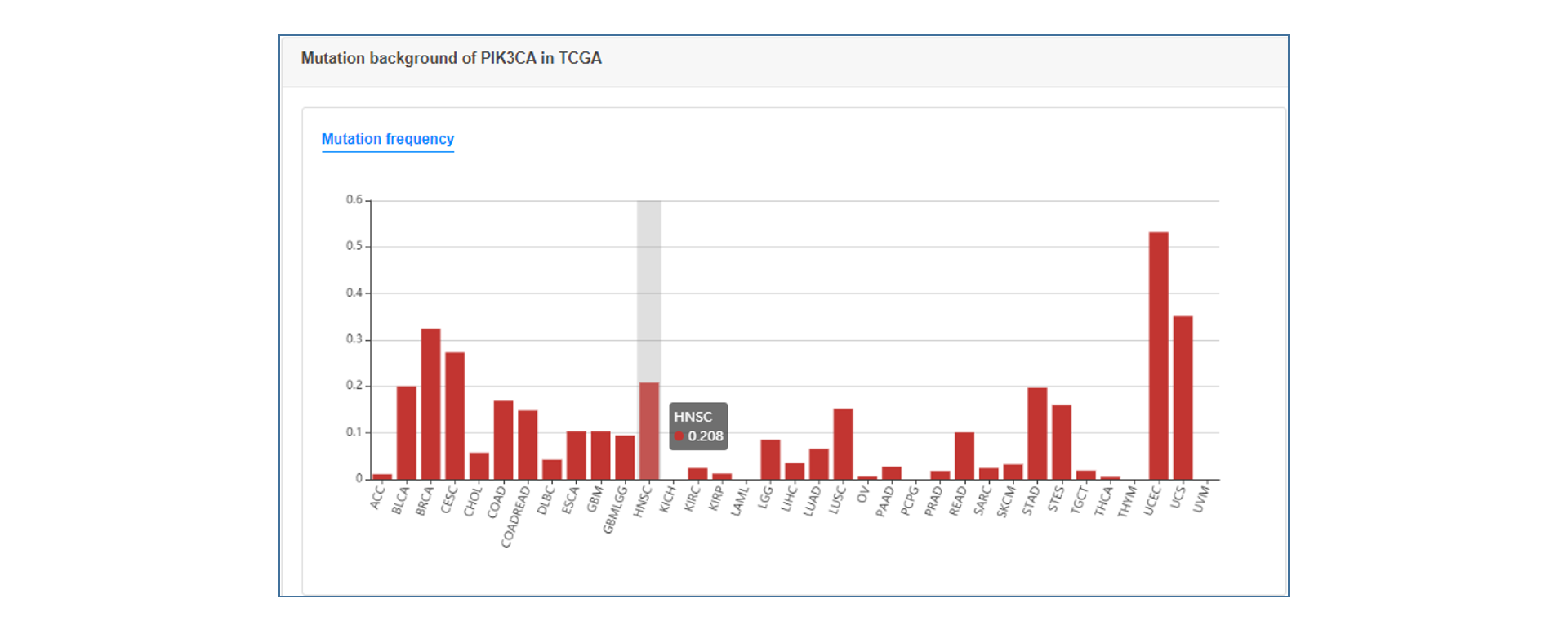
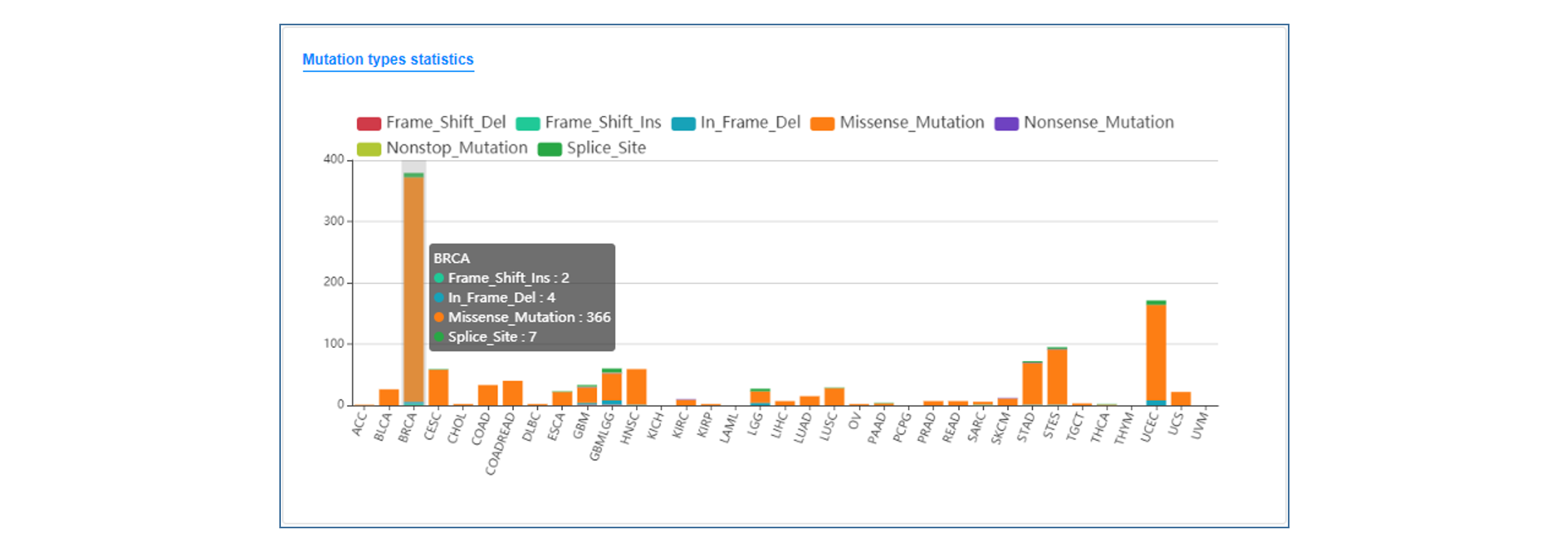
Clinical evidences grading
In DrugCVar, the clinical evidences of drugs were devided into ten levels, the details are shown below.

